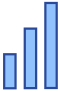
- increased red blood cell distribution width / IMPC
B6;129P2-Il6ratm1Saj/H
| Status | Available to order |
| EMMA ID | EM:14665 |
| Citation information | RRID:IMSR_EM:14665 Research Resource Identifiers (RRID) are persistent unique ID numbers assigned to help researchers cite key resources (e.g. antibodies, model organisms and software projects) in the biomedical literature to improve transparency and reproducibility in research. See https://www.rrids.org/ for more information. |
| International strain name | B6;129P2-Il6ratm1Saj/H |
| Alternative name | Il6ra (CD126)-/- |
| Strain type | Targeted Mutant Strains : Knock-out |
| Allele/Transgene symbol | Il6ratm1Saj |
| Gene/Transgene symbol | Il6ra |
Information from provider
| Provider | Simon Jones |
| Provider affiliation | School of Medicine, Cardiff University |
| Genetic information | Interleukin 6 receptor alpha (Il6ra or CD126)-deficient mice were generated using a conventional replacement vector (neomycin cassette) to disrupt exons 4, 5, and 6, which encode the structural regions of the receptor important for IL6 recognition. |
| Phenotypic information | Homozygous:Comprehensive phenotypic studies of Il6ra -/- mice have not been performed as yet, only limited immunophenotyping including: 1) CD4+ T cells were nonresponsive to IL6 itself and showed no increase in STAT1 or STAT3 phosphorylation following direct stimulation with IL6 2) TGF-beta 1 failed to induce Th17 development in splenic T cells. Heterozygous:Comprehensive phenotypic studies have not been performed as yet. |
| Breeding history | The targeted clones were injected into C57BL/6J-derived blastocysts. Male chimeras were crossed with C57BL/6J females to produce heterozygous offspring, which were subsequently bred with C57BL/6J mice. Heterozygous mice were intercrossed to create a Il6ra -/- generation [C57BL/6J x 129/Ola] before being bred onto a C57BL/6 background. |
| References |
|
| Homozygous fertile | yes |
| Homozygous viable | yes |
| Homozygous matings required | no |
| Immunocompromised | yes |
Information from EMMA
| Archiving centre | Mary Lyon Centre at MRC Harwell, Oxford, United Kingdom |
Disease and phenotype information
IMPC phenotypes (gene matching)
MGI phenotypes (gene matching)
- abnormal inflammatory response / MGI
- liver inflammation / MGI
- no abnormal phenotype detected / MGI
- decreased tumor growth/size / MGI
- increased hepatocyte apoptosis / MGI
- decreased hepatocyte proliferation / MGI
- decreased incidence of tumors by chemical induction / MGI
- abnormal wound healing / MGI
- decreased susceptibility to parasitic infection / MGI
- increased circulating aspartate transaminase level / MGI
- growth/size/body region phenotype / MGI
- abnormal CD4-positive, alpha-beta T cell physiology / MGI
- decreased circulating tumor necrosis factor level / MGI
- decreased circulating interleukin-1 beta level / MGI
- abnormal circulating serum amyloid protein level / MGI
- decreased susceptibility to parasitic infection induced morbidity/mortality / MGI
Literature references
- Loss of CD4+ T cell IL-6R expression during inflammation underlines a role for IL-6 trans signaling in the local maintenance of Th17 cells.;Jones Gareth W, McLoughlin Rachel M, Hammond Victoria J, Parker Clare R, Williams John D, Malhotra Raj, Scheller Jürgen, Williams Anwen S, Rose-John Stefan, Topley Nicholas, Jones Simon A, ;2010;Journal of immunology (Baltimore, Md. : 1950);184;2130-9; 20083667
Information on how we integrate external resources can be found here
INFRAFRONTIER® and European Mouse Mutant Archive - EMMA® are registered trademarks at the European Union Intellectual Property Office (EUIPO).